From trio information to full families
Emil M. Pedersen
2025-11-17
Source:vignettes/FromTrioToFamilies.Rmd
FromTrioToFamilies.RmdIn this document, we will present how we can go from trio information
to full families that can be used to calculate kinship matrices. By trio
information, we specifically mean knowing the id of the child and the id
of the child’s mother and father. Kinship matrices are essential when
estimating the liabilities with the estimate_liability()
function of the package. This addition help with the process of
identifying related individuals and subsequent construction of the
kinship matrix.
From trio information to graph
The trio information can be used to create extended families manually by first identifying parents, grandparents, great-grandparents, etc.. From there, siblings, aunts and uncles, cousins, etc.. can also be identified. However, this is a tedious process and it is easy to miss family members. We have developed a function that can find all family member that are related of degree or closer that does not rely on the tedious process of identifying each family role manually.
Below is an example data set of a family. It contains half-siblings, half-aunts and -uncles, as well as cousins and individuals that have married into the family. An example is mgm meaning maternal grandmother, hspaunt meaning paternal half-aunt, or hsmuncleW meaning maternal half-uncle’s wife. We construct the example dataset with tribble because it enables row-wise construction, which mirrors how trio information is typically stored and helps readability in small example datasets.
# Setting seed:
set.seed(555)
# constructing family (trio) data
family = tribble(
~id, ~momcol, ~dadcol,
"pid", "mom", "dad",
"sib", "mom", "dad",
"mhs", "mom", "dad2",
"phs", "mom2", "dad",
"mom", "mgm", "mgf",
"dad", "pgm", "pgf",
"dad2", "pgm2", "pgf2",
"paunt", "pgm", "pgf",
"pacousin", "paunt", "pauntH",
"hspaunt", "pgm", "newpgf",
"hspacousin", "hspaunt", "hspauntH",
"puncle", "pgm", "pgf",
"pucousin", "puncleW", "puncle",
"maunt", "mgm", "mgf",
"macousin", "maunt", "mauntH",
"hsmuncle", "newmgm", "mgf",
"hsmucousin", "hsmuncleW", "hsmuncle"
)
# simulating sex status on ambiguous individuals
thrs = tibble(
id = family %>% select(1:3) %>% unlist() %>% unique(),
sex = case_when(
id %in% family$momcol ~ "F",
id %in% family$dadcol ~ "M",
TRUE ~ NA)) %>%
mutate(sex = sapply(sex, function(x) ifelse(is.na(x), sample(c("M", "F"), 1), x)))The object family is meant to represent the trio
information that can be found in registers. It is possible to have
multiple families in the same input data or single individuals with no
family links.
graph = prepare_graph(.tbl = family,
node_attributes = thrs,
fcol = "dadcol",
mcol = "momcol",
icol = "id")
graph## IGRAPH 08de485 DN-- 31 44 --
## + attr: name (v/c), sex (v/c)
## + edges from 08de485 (vertex names):
## [1] dad ->pid mom ->pid dad ->sib
## [4] mom ->sib dad2 ->mhs mom ->mhs
## [7] dad ->phs mom2 ->phs mgf ->mom
## [10] mgm ->mom pgf ->dad pgm ->dad
## [13] pgf2 ->dad2 pgm2 ->dad2 pgf ->paunt
## [16] pgm ->paunt pauntH ->pacousin paunt ->pacousin
## [19] newpgf ->hspaunt pgm ->hspaunt hspauntH->hspacousin
## [22] hspaunt ->hspacousin pgf ->puncle pgm ->puncle
## + ... omitted several edgesThe object graph is a directed graph constructed from
the trio information in family and is build using the
igraph package. The direction in the graph is
from parent to offspring.
From graph to subgraph and kinship matrix
We can construct a kinship matrix from all family members present in
family, or we can consider only the family members that are
of degree
.
We can identify the family members of degree
like this:
# get_family_graphs wraps make_ego_graph returns a formatted tbl
fam_graph = get_family_graphs(pop_graph = graph,
ndegree = 2,
proband_vec = V(graph)$name)
fam_graph## # A tibble: 31 × 2
## fid fam_graph
## <chr> <list>
## 1 pid <igraph>
## 2 sib <igraph>
## 3 mhs <igraph>
## 4 phs <igraph>
## 5 mom <igraph>
## 6 dad <igraph>
## 7 dad2 <igraph>
## 8 paunt <igraph>
## 9 pacousin <igraph>
## 10 hspaunt <igraph>
## # ℹ 21 more rowsfam_graph is a tibble with one row per proband (here all
individuals in the graph are probands). The first column is the family
ID, typically the name of the proband the family graph is centred
around, and a column fam_graph containing the corresponding
family graphs as igraph objects. We can plot one of the identified
family graphs with the standard plot function from the igraph package,
however it is not ideal for pedigrees past a certain size.
plot(fam_graph$fam_graph[[1]], # choosing pid's family graph
layout = layout_as_tree,
vertex.size = 27.5,
vertex.shape = "rectangle",
vertex.label.cex = .75,
edge.arrow.size = .3) 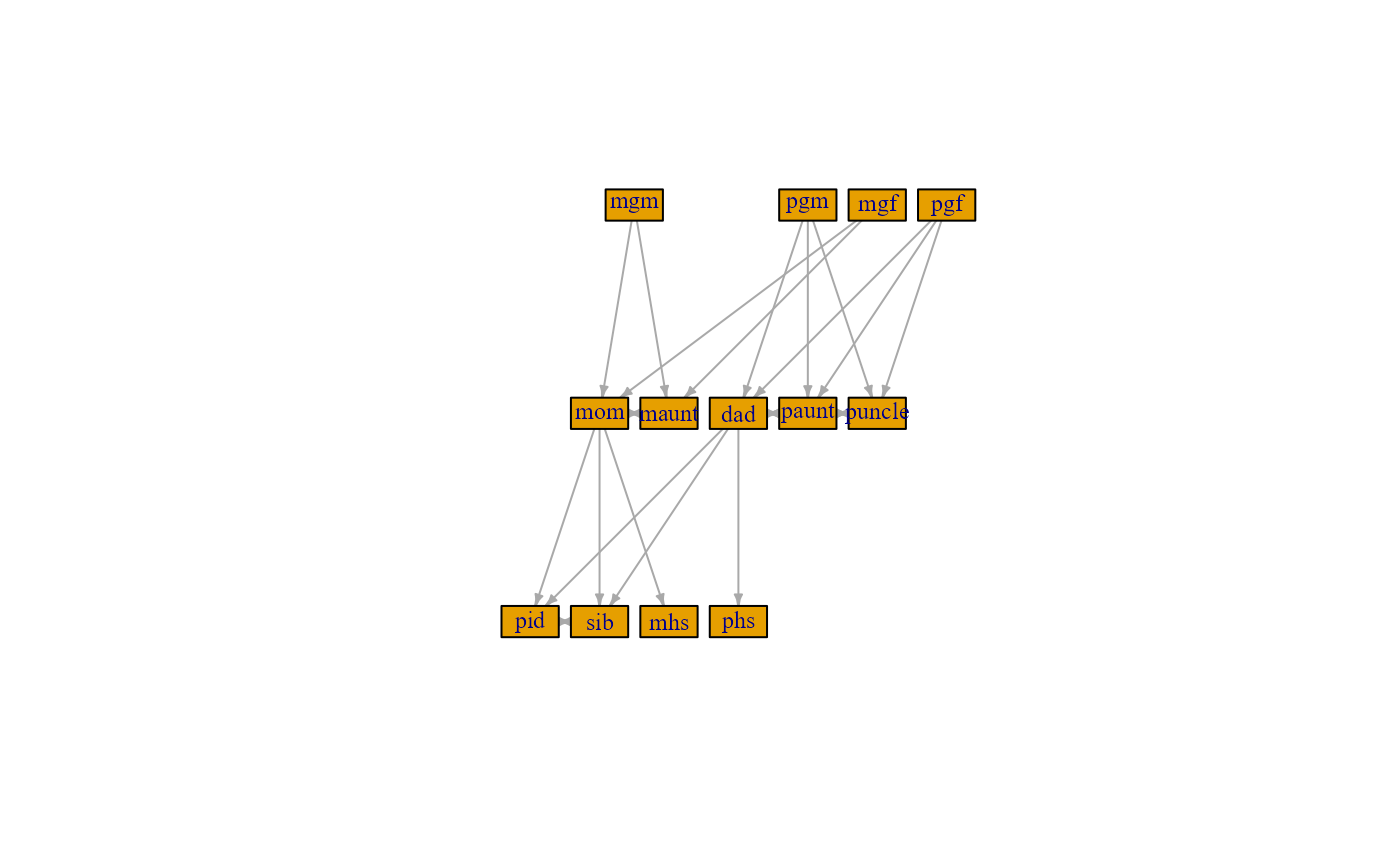
In particular, individuals such as paternal uncle’s child (i.e a cousin, coded as pucousin above) is not present with this relatedness cut-off as such family members are of degree .
Calculate kinship matrix
Finally, the kinship matrix can be calculated with
get_kinship() in the following way:
get_kinship(fam_graph$fam_graph[[1]], h2 = 1, index_id = "pid", add_ind = FALSE)## pid sib mhs phs mom dad paunt puncle maunt mgm pgm mgf pgf
## pid 1.00 0.50 0.25 0.25 0.5 0.5 0.25 0.25 0.25 0.25 0.25 0.25 0.25
## sib 0.50 1.00 0.25 0.25 0.5 0.5 0.25 0.25 0.25 0.25 0.25 0.25 0.25
## mhs 0.25 0.25 1.00 0.00 0.5 0.0 0.00 0.00 0.25 0.25 0.00 0.25 0.00
## phs 0.25 0.25 0.00 1.00 0.0 0.5 0.25 0.25 0.00 0.00 0.25 0.00 0.25
## mom 0.50 0.50 0.50 0.00 1.0 0.0 0.00 0.00 0.50 0.50 0.00 0.50 0.00
## dad 0.50 0.50 0.00 0.50 0.0 1.0 0.50 0.50 0.00 0.00 0.50 0.00 0.50
## paunt 0.25 0.25 0.00 0.25 0.0 0.5 1.00 0.50 0.00 0.00 0.50 0.00 0.50
## puncle 0.25 0.25 0.00 0.25 0.0 0.5 0.50 1.00 0.00 0.00 0.50 0.00 0.50
## maunt 0.25 0.25 0.25 0.00 0.5 0.0 0.00 0.00 1.00 0.50 0.00 0.50 0.00
## mgm 0.25 0.25 0.25 0.00 0.5 0.0 0.00 0.00 0.50 1.00 0.00 0.00 0.00
## pgm 0.25 0.25 0.00 0.25 0.0 0.5 0.50 0.50 0.00 0.00 1.00 0.00 0.00
## mgf 0.25 0.25 0.25 0.00 0.5 0.0 0.00 0.00 0.50 0.00 0.00 1.00 0.00
## pgf 0.25 0.25 0.00 0.25 0.0 0.5 0.50 0.50 0.00 0.00 0.00 0.00 1.00A function called graph_to_trio() has been included in
the package, which can convert from the graph object back into a trio
object. This function is useful if you want to use the functionality of
other packages that rely on trio information. One such example is using
the plotting functionality of pedigrees in kinship2.
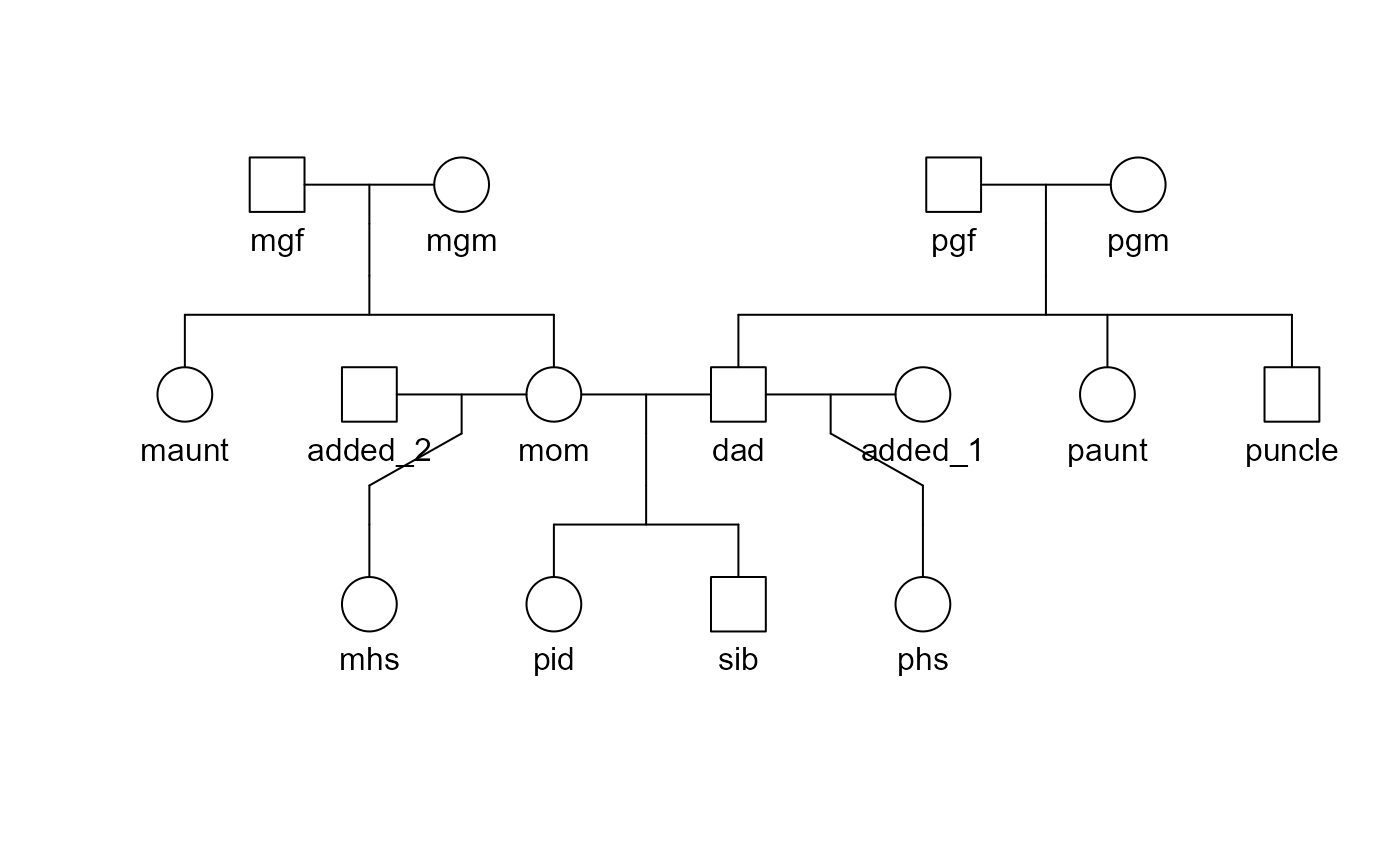
trio = graph_to_trio(graph = fam_graph$fam_graph[[1]], fixParents = TRUE)
trio## # A tibble: 15 × 4
## id momid dadid sex
## <chr> <chr> <chr> <chr>
## 1 pid "mom" "dad" F
## 2 sib "mom" "dad" M
## 3 mhs "mom" "added_2" F
## 4 phs "added_1" "dad" F
## 5 mom "mgm" "mgf" F
## 6 maunt "mgm" "mgf" F
## 7 dad "pgm" "pgf" M
## 8 paunt "pgm" "pgf" F
## 9 puncle "pgm" "pgf" M
## 10 mgf "" "" M
## 11 pgf "" "" M
## 12 mgm "" "" F
## 13 pgm "" "" F
## 14 added_1 "" "" F
## 15 added_2 "" "" Mwhich can be used to utilise the powerful plotting tool kit available in the kinship2 package.
pedigree = with(trio,kinship2::pedigree(id = id, dadid = dadid,momid = momid,sex = sex))
plot(pedigree)